The COVID-19 pandemic does not appear to go away any time soon. It has become an unchanging part of everyday life, at least for the foreseeable future. However, the SARS-CoV-2 virus has been busy mutating into numerous variants. Many of these mutants—particularly variants of concern (VOCs)—are more lethal than the original virus and can evade antibodies. Now, scientists have created a global 'atlas' that illustrates how over 150 antibodies target the notorious spike protein of the coronavirus and its variants.
In the study published in the journal Cell, researchers demonstrated that 152 different antibodies attacked regions of the spike. The authors ascertained that some of these antibodies could effectively neutralize the newer variants, and identified regions of the spike that have developed resistance to antibody attacks.
"Emerging data show that vaccines still confer some protection from new SARS-CoV-2 variants, and our study shows how that works from an antibody standpoint. These data can help us think about what the best kind of booster vaccine might be by studying how the repertoire of human antibodies recognizes the spike protein," said Dr. Duane Wesemann, corresponding author of the study in a statement.
Spike Mutations and Variants
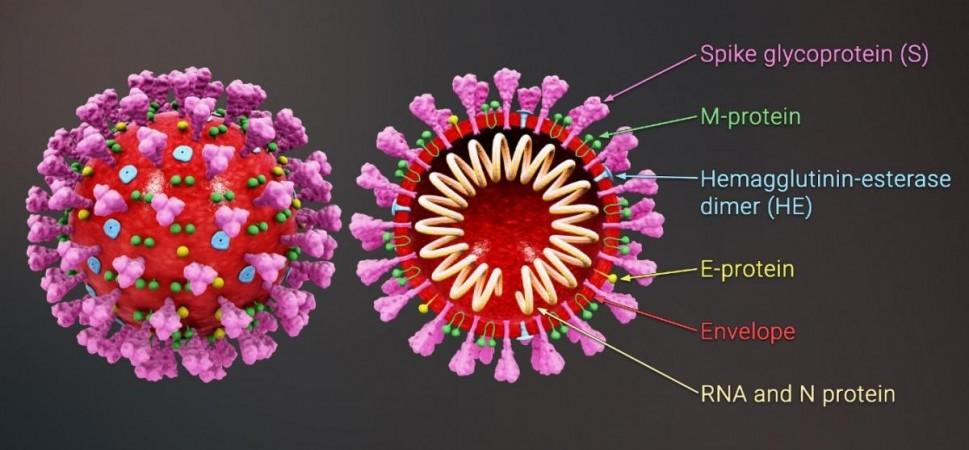
It is natural for viruses such as the SARS-CoV-2 to evolve as they make their way into different populations. While most of the mutations have no significant impact on the nature of the virus, some can equip them with potent abilities. Most of the mutations in the newer variants such as VOCs are located on the 'S- protein' or 'spike' of the SARS-CoV-2. The spike is a hook-like structure present on the surface of the virus that it uses to invade cells. It is divided into two parts—S1 subunit and S2 subunit.
The S1 subunit comprises of N-terminal domain (NTD) and a receptor-binding domain (RBD). Fusion peptide (FP), transmembrane (TM) domain, cytoplasmic domains (CD), and heptapeptide repeat sequence 1 (HR1) and heptapeptide repeat sequence 2 (HR2) form the S2 subunit. These individual regions consist of their own unique units known as residues.
Immunoevasion (the ability to evade antibodies acquired through infection or vaccination) and higher infectivity and increased transmissibility are some of the weapons that spike mutation provide the variants. Currently, Currently, four variants of SARS-CoV-2 have been declared as VOCs by the WHO—Alpha (B.1.1.7- UK), Beta (B.1.351- South Africa), Delta (B.1.617.2- India), and Gamma (P.1- Brazil)
Looking for 'Footprints'
For the study, the authors investigated the Memory B cells of 19 adults who had contracted COVID-19 in March 2020, much before the new variants emerged. Memory B cells are a type of white blood cells that are an integral part of the adaptive immune system. They are responsible for the production of antibodies against invading pathogens. The team examined how these antibodies, along with those described by them, bound with the spike protein models of the Alpha, Beta, and Gamma variants. Currently, a study of the Delta variant is in progress.
On the whole, it was confirmed that seven major 'footprints' on the spike protein were the areas where hundreds of antibodies that were being analyzed bind. While several of these antibodies 'competed' with each other to bind to the same particular regions on the spike of the original SARS-CoV-2 virus, some of them lost their lethality when it came to the newer strains. Some were broadly responsive neutralizers.
Neutralization Ability of Antibodies

In the earlier versions of the SARS-CoV-2 spike, antibodies that bind with two specific regions called RBD-2 and NTD-1 served as the strongest neutralizers. Among the variants, the spike of the Beta variant displayed the largest evasive abilities against existing antibodies; particularly most of the antibodies that bind with RBD-2 and NTD-1 sites. Interestingly, some antibodies that bind with another region known as S2-1 were able to recognize spikes of other related coronaviruses such as SARS-CoV-1, MERS-CoV, and the common cold.
Dr. Wesemann explained that the making of distinct antibodies that vie for one particular region of the coronavirus lets the immune system be adaptable. "Otherwise-redundant recognition by antibodies targeting the same footprint of one version of the virus confers recognition depth of the same footprint on variants, and some antibodies maintain high neutralization potency against all the variants. Now that we can identify the antibodies that are more broadly reactive to all of the variants, we can think about how to elicit them more strongly in a vaccine," he illustrated.















